
The Tower of Babel by Pieter Bruegel the Elder (1563)
Trends in Bioinformatics Programming Languages on GitHub
Bioinformatics as a discipline came into its own with Next Generation Sequencing (NGS) technology. Widespread use of NGS technology created vast quantities of DNA sequencing data which drove development of tools for analysis of the data. Many of these tools were written using programming languages familiar to academic researchers, often Python or R, which, though fast in the writing, are slow in the running. Given the vast quantity of data that typically need to be processed, the lack of performance of these languages can limit the possible analysis. As a result, a number of new programming languages, such as Rust, Julia, Go, or Nim, have been noticed by the bioinformatics community to help improve performance. Researchers at the Massachusetts Institute of Technology have developed a new language, Seq, in 2019 to improve performance while retaining the productivity of Python.
Programming language for bioinformatics matters in terms of balancing productivity and running times. But what programming languages are people actually using for bioinformatics? How has language use changed over time in this field? What new languages are gaining traction?
Pamela Russell, and co-authors, provide a partial answer to the language choices people are making. They analyze the relationship between code properties, development activity, developer communities, and software impact. The authors note the vast majority of code repositories reported in the Oxford Academic journal Bioinformatics, an important journal in the discipline, are hosted on GitHub (Figure 1). This is an important result, since GitHub can then be used to gather reliable data on programming language use for bioinformatics.
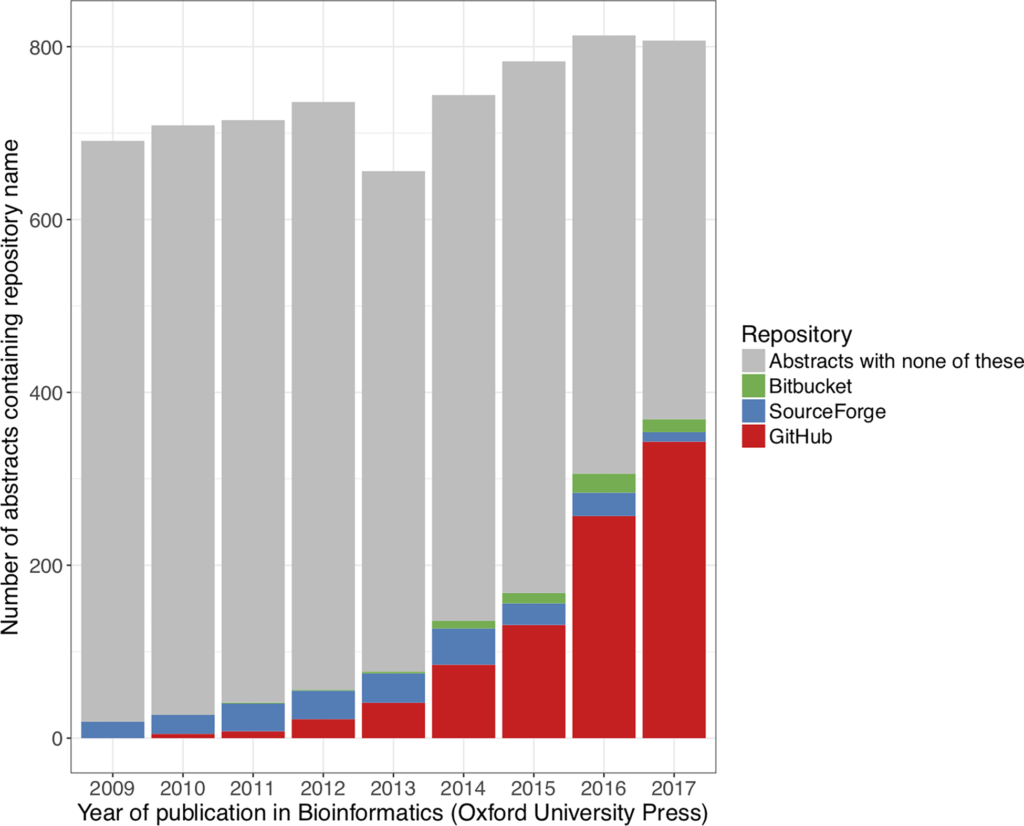
https://doi.org/10.1371/journal.pone.0205898.g001
The authors then consider languages included in at least 50 main repositories and report lines of code per file as a function of number of files for each language shown below (Figure 2). The results show a prevalence of Bourne Shell, C, C++, Perl, Python, and R used between 2009 and 2017.
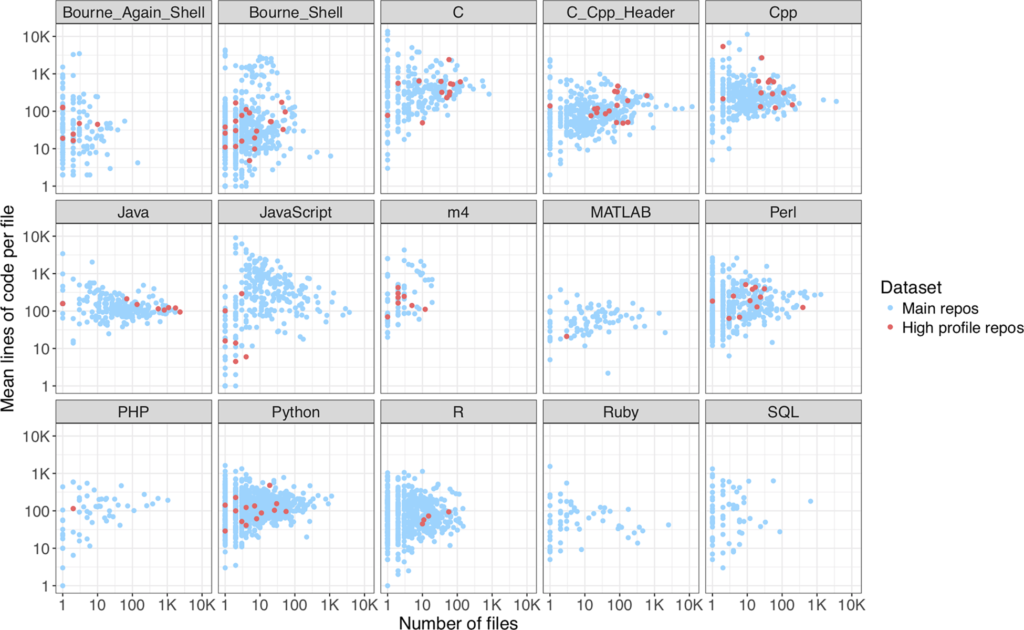
https://doi.org/10.1371/journal.pone.0205898.g003
While this partial answer is helpful, it does not show change over time and thus, cannot be informative about language trends or new languages gaining traction. To answer these questions, I returned to the Bioinformatics website and noticed the search term “github.com” returned a page of results that provided the GitHub URL of the repository reported in the article. And so, I wrote a Python (yes, of course) script using Selenium to pull down all the search results, Beautiful Soup to parse the results, git to clone the repositories (2111 in all) and cloc to count lines of code. The result is a JSON file containing the GitHub URL and a list of languages used in each repository in decreasing lines of code order. I then counted the number of times one of the top three languages (by lines of code) appeared in each repository, a number I termed the incidence of that language in the repository, by year. Sorting the resulting pandas DataFrame by total incidence produced, with the lovely heatmap capabilities of Matplotlib, the top 15 languages are shown in Figure 3, which also shows their corresponding relative incidence, relative to the total for the year, for each year in the past decade.
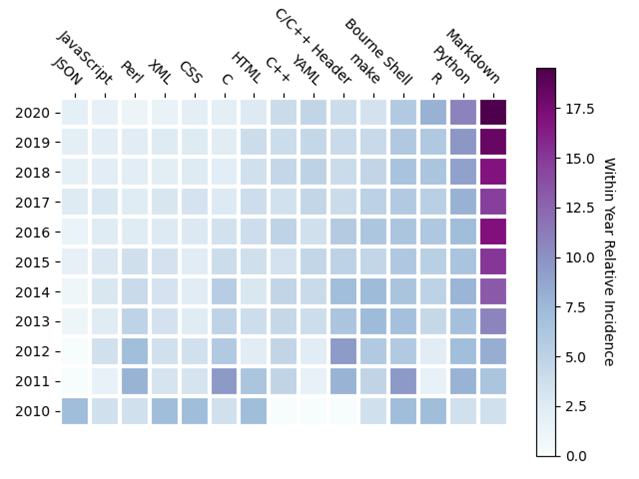
I was surprised but heartened, to see Markdown dominating the within year incidence. While not a programming language, I interpreted this result to indicate the importance of documentation, in a standard format, has been noted by the bioinformatics community. I was not surprised to see Python and R as the top two programming languages, though interested to note the within year relative incidence of each has been relatively constant over the last ten years. C and C++ are in the list, but further down, with C++ gaining over C. To see the languages gaining traction, I computed the change in incidence from 2019 to 2020, which I call the 2020 year-on-year incidence increase. Sorting the same resulting DataFrame by this 2020 year-on-year incidence increase produces the top 15 languages shown in Figure 4, which also shows their corresponding relative incidence for each year in the past decade, as before.
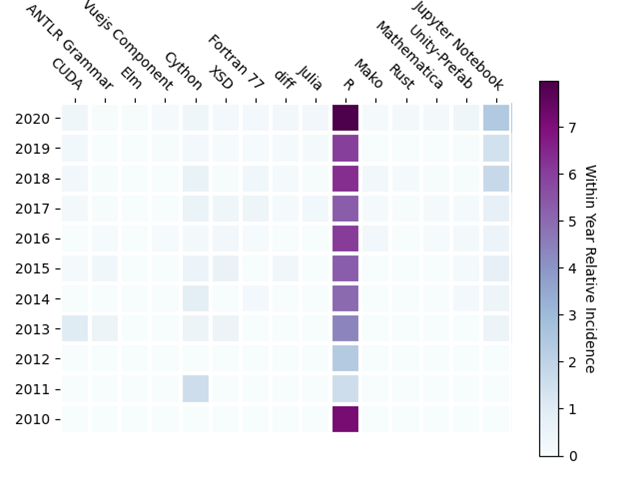
Since the within year relative incidence is small in this set of entries, the year-on-year incidence increase is noisy. As a result, some unexpected entries appear, among them Unity-Prefab or Fortran 77. More interesting, and relevant, are the increasing adoption of the conventional programming language R, and the new programming languages Rust and Julia. To bring out the differences in relative incidence of these lightly used entries, the results in Figure 4 are shown in Figure 5 with R removed.
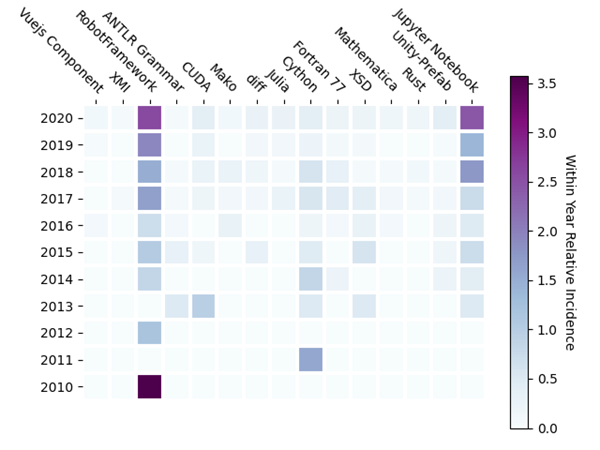
Jupyter Notebook (a web application that allows people to create and share code, equations, visualizations, and text) becomes prominent, steadily gaining incidence each year. Since Jupyter supports Python and R, perhaps this should be expected. However, Jupyter also supports Julia, and Scala, and many other languages, so it would be interesting to know which languages are represented in this Jupyter notebook use. The appearance of Robot Framework (used for test and robotic process automation) was a surprise to me. Though not among the top 10 entries with the largest year on year incidence increase, Robot Framework shows steady incidence growth over the last decade. I suspect this result is localized to a very specific use case.
From these analyses, here is what I’ve learned about language use in Bioinformatics:
- Python and R each capture a steady 10% or so of the bioinformatics programming language use.
- R has gained more rapidly than Python over the last decade.
- C and C++ are the only programming languages in the top 20by incidence used for run time performance.
- Rust and Julia are the only new programming languages in the top 20 by 2020 year on year incidence increase.
Stay tuned! In later posts on this topic I plan to investigate code quality, use of pipeline frameworks, and to track the adoption, or not, of the newer programming languages.

