Transfer Learning for Classification in Ultra-small Biomedical Datasets, Part II
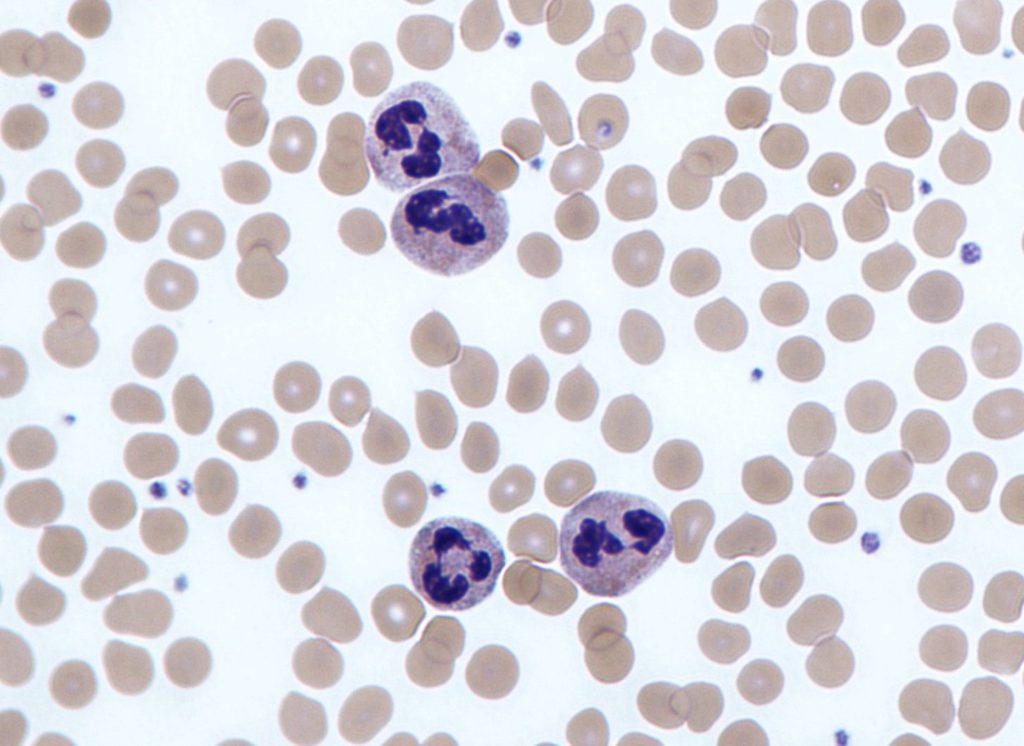
Four neutrophils amongst red blood cells (image by Dr Graham Beards / CC BY-SA)
How many purple neutrophil white blood cells are in the image above? Such a question isn’t just a toy example — counting the number and ratio of various types of blood cells is an informative metric for patient diagnosis and care. If you answered, “four,” you’re correct — although if you didn’t know what a neutrophil was and answered “twenty,” I’d take that too (there are twenty purple blobs in the image). What if the image was this one:
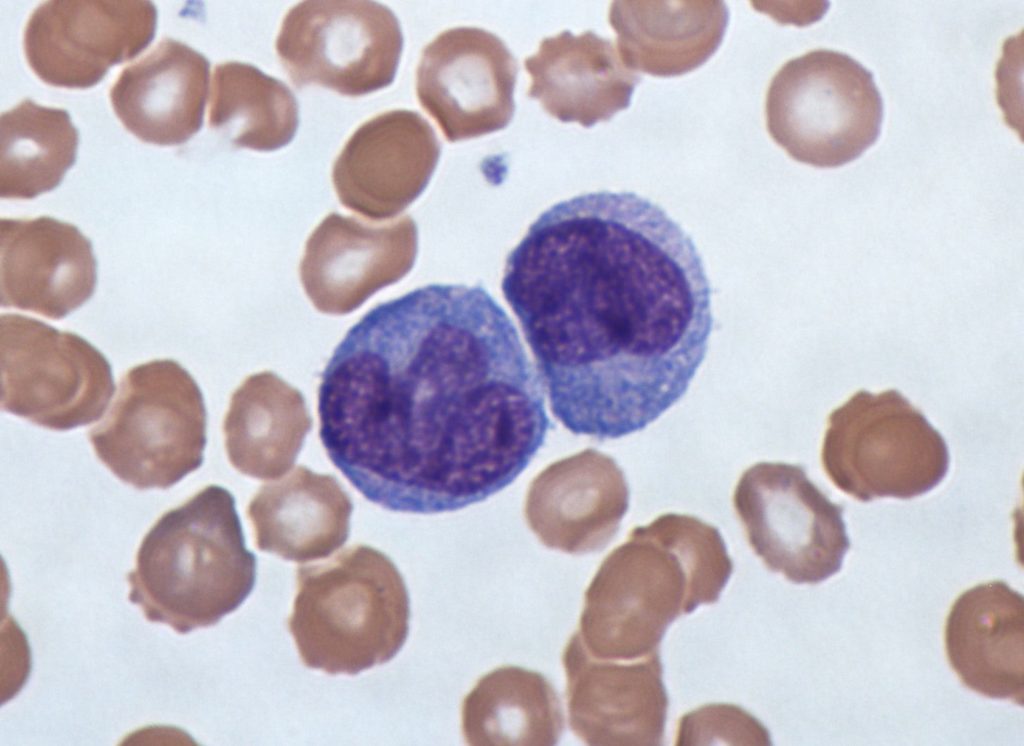
It turns out, there are no neutrophils in this image — the two large purple cells are another type of blood cell: monocytes. And while it’s still somewhat easy for untrained humans to detect differences between neutrophils and monocytes, it becomes harder for computer vision models to do the same, especially when these cells are within smears that contain red blood cells, as above. Therefore, automated approaches to cell counting and individual cell classification will often rely on segmentation of the cells of interest before attempting to identify them.
In our previous blog post about transfer learning using biomedical imagery, we explored how classifying cell cultures, made up of many cells, is a computer vision task that seems to benefit from transfer learning from models that were previously trained on ImageNet, before being applied to our datasets. These datasets can be thought of as texture-based challenges, where the textures are the different cell shapes across different classes. However, biomedical applications are often interested in analyzing individual, segmented cells. Our hypothesis is that transfer learning from ImageNet is not as likely to aid these models, compared to the textured datasets above, because it’s even less clear how to recycle lower-level features into higher-order ones that ImageNet-based models have learned.
Segmented Cell Classification
Previously, we compared pre-trained vgg19 and ResNet18, or rather just the lower layers of these two models, against a deep and shallow CNN to see how helpful transfer learning might be. The details of this experimental setup can be found in our previous blog post; to summarize, we used cross-validation across twenty trials to measure each model’s performance on the same holdout per dataset. In this post, we’re going to now compare these various models across datasets with segmented cells.
Our first dataset is of synthetically generated cells, where the nuclei (in red), cell bodies (in blue), and various proteins (in green) were simulated to mimic real biological structures, and could be used to train models that would later predict proteins from real images. We resized each 1024×1024 pixel image to 224×224 to avoid losing cell structures, and trained the model to distinguish between the six protein patterns below:
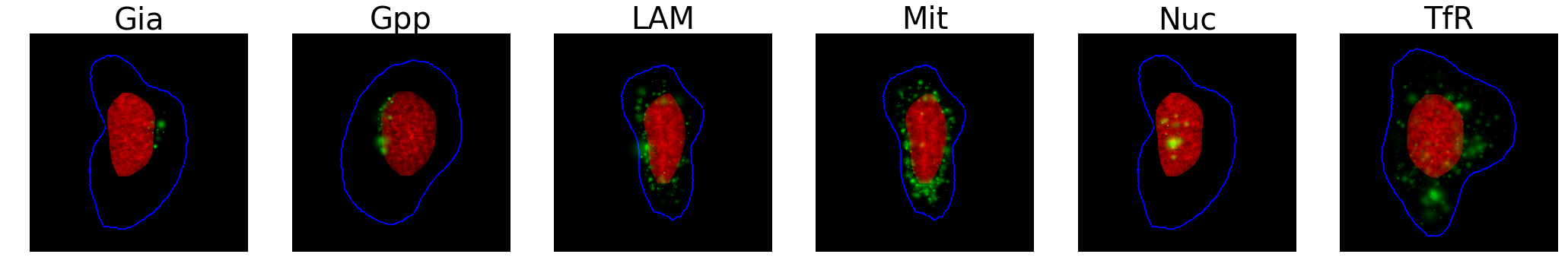
Unlike the previous benchmarks, there wasn’t really a difference between ImageNet-based modelsand a custom CNN. Voting also generated more accurate models, again, but did seem to yield superior results when using just the lower layers with transfer learning.
We also investigated the performance of these architectures on another cell segmentation problem, where red blood cells needed to be labeled as infected with malaria or healthy, below. These images ranged from around 150×150 to 60×60 pixels, which we resized to 224×224. We hypothesized that our models would perform well, as the problem is easy for humans, and that transfer learning from ImageNet wouldn’t yield much of a benefit since we’re merely looking for purple smears.

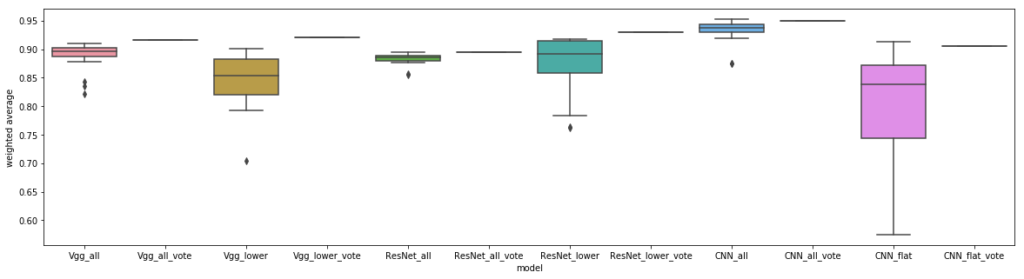
Indeed, overall model performance was strong, with our custom CNN proving slightly more effective. We hypothesize that these individual cells were quite dissimilar from ImageNet, but otherwise simple to classify, which is why transfer learning did not help. Surprisingly, our un-tuned CNN seemed to perform just about as well as a paper that build models off the entire dataset (so ~28,000 images, not our subset of 1000, but evaluated similarly with cross-validation and holdout), although perhaps this result is not that surprising after all, given that smaller datasets may only knock off a few percentage points in accuracy.
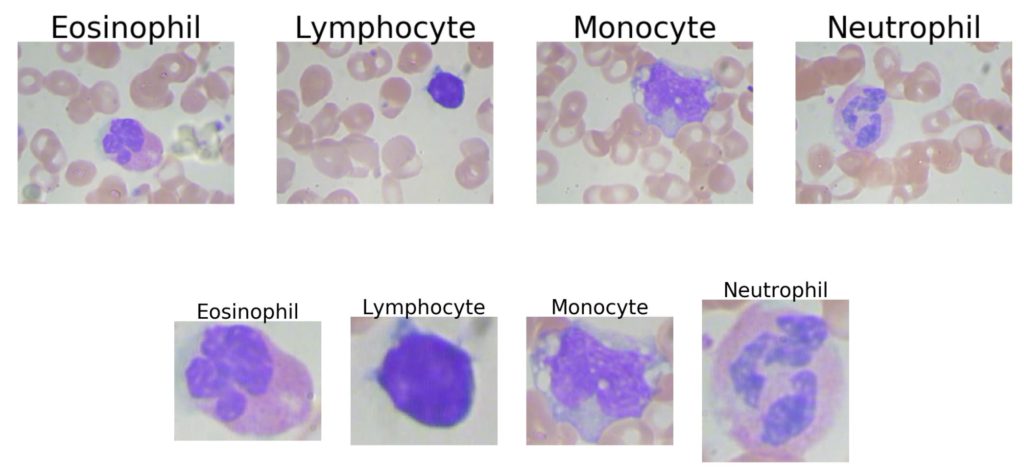
Both these segmentation datasets had at least one hundred high-quality images to train from for each class, and classes were balanced. We were curious how a model might perform that had severe class imbalances, with close to the same number of average images per class. The blood cell dataset below labels each image with the type of the white blood cell that is stained in purple; the surrounding tan cells are red blood cells. The original dataset contains 640×480 pixel images, but we also experimented with manually segmenting the white blood cells of interest, yielding images that were around 200×200 pixels. For both trials, we resized the images to be 224×224 pixels and trained with a learning rate of 1e-2 and 1e-3, respectively.
We predicted that it would be difficult to identify white blood cell types in the presence of red blood cells, as the latter adds clutter and could confuse the models. As for the segmented cells, although the eosinophils and neutrophils looked very similar to the untrained eye, the class imbalance was in their favor. Overall, this problem is difficult for naive humans, and given the limited number of images, the clutter of red blood cells, and the blurriness of even the segmented cells, we did not expect these models to perform as well as some of our earlier benchmarks.
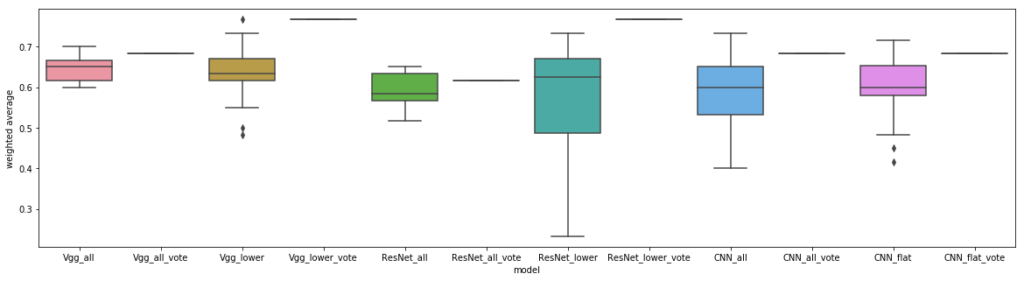
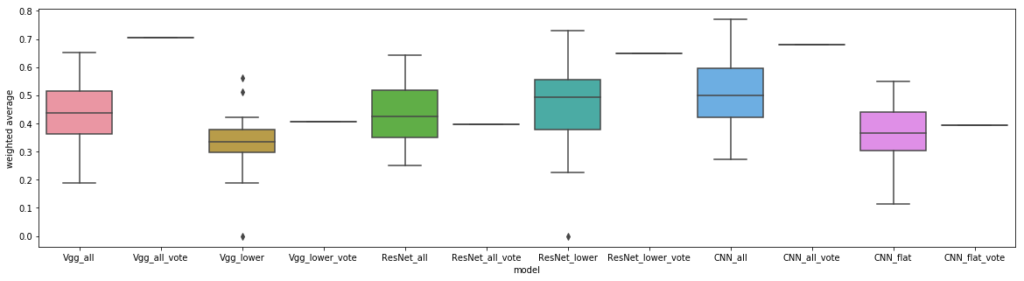
Our results below show that transfer learning from ImageNet based models may hurt with this classification, even for the segmented cells. For the first time, ResNet outperformed vgg, though still coming in second place to our custom CNN. Interestingly, other work has implemented cell segmentation for this challenge, with a 93% classification accuracy, similar to our CNN performance on segmented cells (and they also used a CNN to generate their features).
Other Biomedical Image Datasets
The datasets above ranged from moderate, to easy, for humans to classify, and there was generally a strong correlation between ease of human judgment, and model performance. What if we try to build models that try to solve a classification task that’s trivial for humans, but hard for machines (without added help)? We were curious if we could even build a useful model to count a handful of feline reticulocyte cells in an individual image. Although actually segmenting and counting these cat cells would be a far better approach (which someone else has investigated) if we wanted to truly excel on this dataset, we were curious if transfer learning, and specifically, vgg, would outperform the other models.
For this dataset, we resized the 300×300 pixel images to 224×224, instead of cropping them (to prevent cutting out a cell), and only used images from the original dataset that had the black borders shown below. Labels were counts of how many full blue cells each image contained: one, two, three, four, or five-plus. We predicted that although this is a simple problem for humans, without segmenting the cells, our models would have difficulty making these judgments.
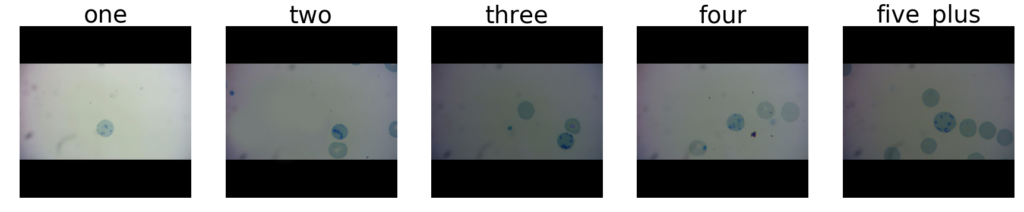
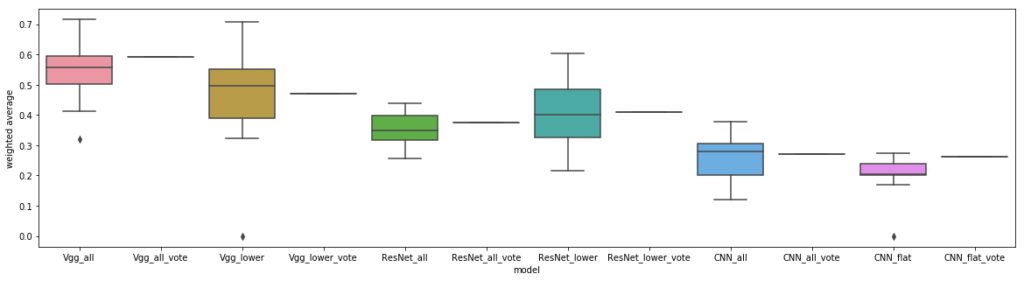
Although the overall classification performance for these models was very low, due to the difficulty of the problem without segmentation, we did observe that the basic vgg model dominated. Out of all our datasets, transfer learning yielded the biggest relative gains on this dataset, and this dataset was also the most difficult one for machines to learn. It makes intuitive sense that for a problem like this, which is easy for humans, but hard for machines (without segmentation), that what can be recycled from transfer learning off of ImageNet might be relevant for the type of aggregation we’d like these models to learn to do, which is to identify cells and count them.
Alternatively, simply normalizing and binarizing the images, and counting the ratio of white versus black pixels may ultimately have been a more accurate approach. We were curious what the differences might be between transfer learning and our regular CNN, so we decided to graph the activations between the two models for a similar intermediate layer, at the end of training:
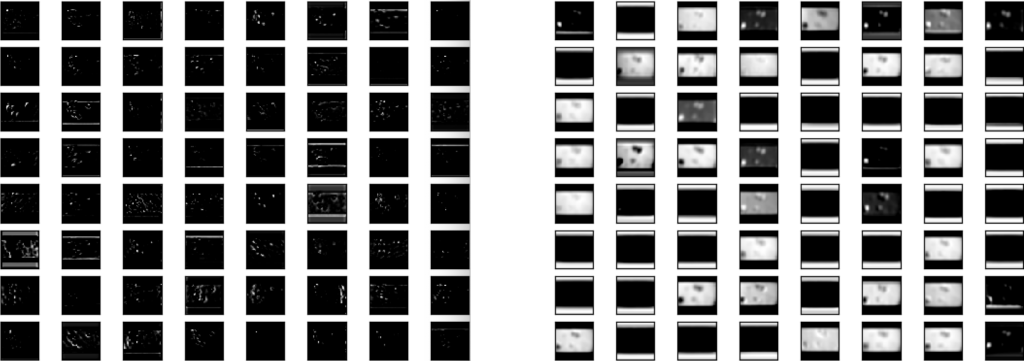
Interestingly, the activations of the poorer-performing CNN (on the right) tend to focus more on the boundaries between the white part and black parts of the original image background, which higher layers of the model may learn are not useful to distinguish between classes. We will investigate better pre-processing in our next blog post.
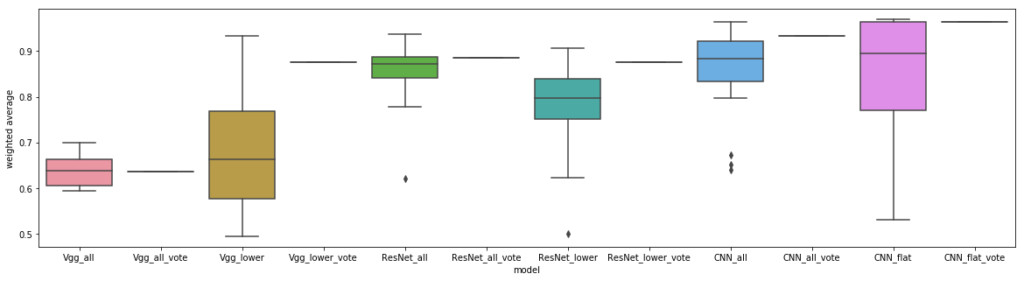
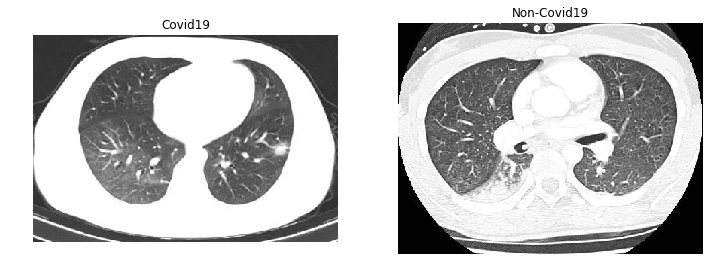
Finally, we had access to another type of common biomedical image: CT scans of large areas of the body, in this case, the lungs of COVID-19 patients versus other patients. Unlike our other datasets, these roughly 400×300 pixel images are not cell-based. However, we hypothesize that we’re still looking for low-level textural differences between the two classes, although the lack of uniformity between lungs and the surrounding organs would once again probably benefit from some segmentation before we tried to classify the lung tissue. Our results are presented below:
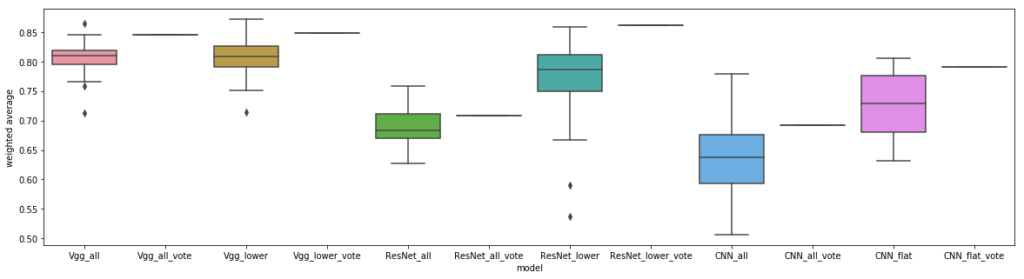
Using the lower layers of vgg works the same as using the whole architecture, which goes against a hypothesis of one of the papers mentioned earlier. We wonder if perhaps a more shallow model is just less prone to overfitting, as we also saw the same trend in our custom CNNs on this dataset. Interestingly, other researchers have also explored using vgg and ResNet to classify Covid19 chest CTs with only a few points higher accuracy, while doing much better on chest X-rays.
Where to Next with Transfer Learning for Ultra-small Biomedical Datasets?
Our original hypothesis that transfer learning would not only help but may even be necessary, to obtain decent performance for these ultra-small biomedical datasets, was supported in about half of these experiments. We didn’t observe conclusive evidence in our experiments that using just the lower layers of transfer learning models would boost performance. Therefore, our new hypothesis is two-fold: 1) that transfer learning is likely to help cell-based biomedical imagery (though not always), and that 2) pre-processing and augmenting raw images may be a useful avenue to explore for increasing model performance. We’ll investigate both of these hypotheses in upcoming blog posts:
- Can we more successfully transfer learn from models trained on large biomedical datasets (as opposed to ImageNet), and,
- What types of edge detection, thresholding, and other transformations can help highlight fine-level textural differences in cell-based images that these models could learn?
Check out our next blog post in this series on ultra-small biomedical dataset classification to find out!
Thanks to my colleagues Felipe Mejia, JohnSpeed Meyers, and Vishal Sandesara
Thank you to the Murphy Lab for making available many of the datasets used here. For further information, see their paper: X. Chen, M. Velliste, S. Weinstein, J.W. Jarvik and R.F. Murphy (2003). Location proteomics — Building subcellular location trees from high resolution 3D fluorescence microscope images of randomly-tagged proteins. Proc. SPIE 4962: 298–306.

