
An Initial Security Assessment of Open Source Bioinformatics Software Packages
Given the growing threat of open source software supply chain attacks, in which attackers compromise open source software packages, we wondered about the security of the open source bioinformatics ecosystem. Bioinformatics packages provide the tools required to analyze genetic sequences and are increasingly important to modern scientific advances. The National Security Commission on Artificial Intelligence has forecasted that advances in biology and bioinformatics will underpin future major scientific breakthroughs related to human health, agriculture, and climate science. The security of bioinformatics software packages is therefore critical not only for U.S. competitiveness but also for national security.
We therefore turned our attention to Bioconda, a popular channel for open source bioinformatics software packages used to store, organize, analyze, visualize, and understand biological sequences. Bioconda provides over 8,000 open source software packages ready to install with Conda, an open source software package management system. Crucially, Bioconda depends on open source contributors to voluntarily add, update, and maintain packages. This means Bioconda users place their trust, even if implicitly, in the people and organizations who write this code.
Our research asked this question: Could defenders detect attackers inserting malicious code into the bioinformatics packages that deal with some of humanity’s most sensitive data? We decided to build a prototype security analysis pipeline to keep attackers out of the bioinformatics software supply chain. Here’s a summary of our findings:
- Our tools did not detect any currently compromised bioinformatics packages.
- There are nonetheless vectors of compromise for these bioinformatics packages.
- Future teams should build security tools that make ecosystem scanning efforts like this easier and less time-consuming.
Those interested in the details should read on. If you’re interested in further work on this topic or collaboration, please contact jmeyers@iqt.org.
An Initial Security Assessment Pipeline for Bioinformatics Packages
We prototyped a set of tools to determine if attackers have already inserted a malicious package or compromised an existing Bioconda package. We hope these tools will benefit the Bioconda maintainers and also companies and researchers with an interest in the security of the bioinformatics software supply chain. This work is part of a larger effort at IQT Labs to address secure code reuse.
Building on the work of a Georgia Tech open source software security dissertation, we created an approach with three components:
- Searching Bioconda recipes and BioContainers Docker files for exfiltration commands.
- Using static analysis to scan Python-based bioinformatics repositories.
- Employing dynamic analysis to identify system calls during Bioconda package installs, BioContainers Dockerfiles builds, and bioinformatics pipeline runs.
We used Dask to distribute the tasks to a cluster. Details and code are available in IQT Labs’ secure-bioinformatics-reuse GitHub repository. We encourage anyone interested in bioinformatics software security to borrow, build on, and provide feedback on this code and approach.
Metadata Analysis
| ssh | sftp | scp | wget | curl | |
|---|---|---|---|---|---|
| Biconda Recipes | 10 | 0 | 5 | 56 | 1442 |
| BioContainers Dockerfiles | 0 | 0 | 2 | 165 | 21 |
Table 1: Occurrence of potential exfiltration commands in Bioconda recipes and BioContainers Dockerfiles
We first searched for commands in Bioconda recipes and BioContainers Dockerfiles that could be used for data exfiltration attacks. Table 1 displays the results.
Inspection of the ssh and scp commands revealed nothing suspicious. The wget commands often obtained software from code repositories like GitHub or sourceforge.net or data from reputable sources such as ftp.ncbi.nih.gov. The results were similar for the wget and curl commands in the BioContainers Docker files except that these commands often obtain code and data from sources other than code repositories. The majority (1,357) of the curl commands in the Bioconda recipes were concentrated in the post-link.sh script for Bioconductor recipes. These curl commands appear to be obtaining data required by the tool. While manual inspection did not reveal anything suspicious, a useful next step would be to evaluate all contacted domains via a threat intelligence service.
Static Analysis
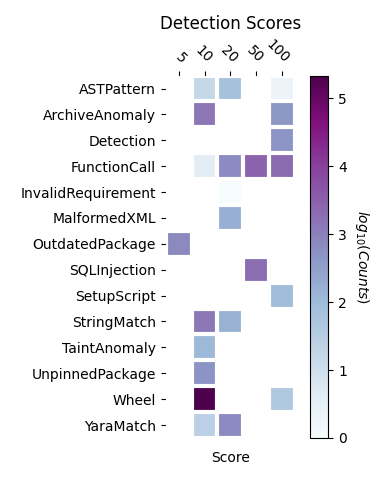
We identified 495 bioinformatics repositories from papers published in the journal Bioinformatics. These repositories were written primarily in Python, enabling us to use the Python static analysis tool Aura. Aura finds indicators (e.g., suspicious code in a setup script) and assigns a score to each indicator. We identified the unique match types (see documentation here) and counted occurrences of each score. The results appear in Figure 1.
Since the scan results are voluminous, we manually inspected a few results focusing on function calls, SQL injections, and anomalous setup scripts. We found no malicious function calls and only a few SQL injection opportunities. Of more concern, we did find opportunities to execute arbitrary Python code during installation of Luigi, and snakePipes, both packages related to building workflows. This vulnerability seems worth noting given the generality of these packages.
Because these repositories were originally included in a prestigious peer-reviewed journal, we suspected it was unlikely that we would find malicious code, and we did not. However, given the limited time available for manual inspection and evident opportunities for attack, future research should run security scans on the long tail of less closely scrutinized bioinformatics packages. We should also mention that assessing the results was time-consuming and our experience highlights the value of projects that make it easier to filter, sort, and view the results of static analysis scans.
Dynamic Analysis
We used strace, a dynamic analysis tool that monitors running code, to identify system calls during the install of 1,028 Bioconda packages, a build of 969 Biocontainer Dockerfiles, and a run of 35 bioinformatics pipelines. We examined the output and concluded that we should focus on executed files and IP addresses. We did not find any executed files of concern.
We did, however, identify 61 IP addresses to which a connection was made during the installations. Figure 2 shows the 20 least and most frequently occurring IP addresses. We focused on the least frequently occurring and public addresses as the most suspicious. However, assessment of the security risk of connections to these IP addresses proved difficult to complete. In some cases, the IP address corresponded to a content delivery network, and so the ultimate endpoint was not readily identified. Future versions of a bioinformatics security pipeline will need a method for assessing IP addresses.
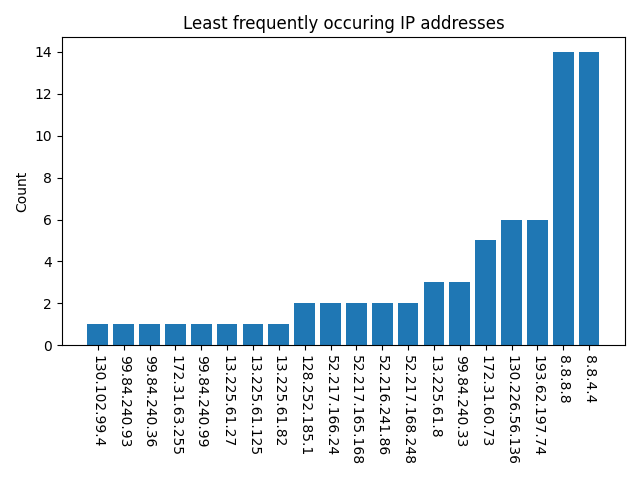
Figure (a) 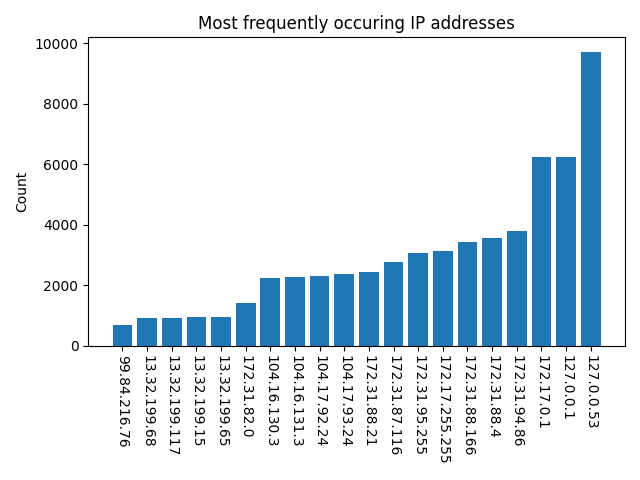
Figure (b)
Trust and Verify: No Malicious Bioinformatics Packages Found…For Now.
What did we learn? Users of bioinformatics packages place their trust, even if implicitly, in the people and organizations that write the code. Simple attacks on the Bioconda build and Conda install process are theoretically possible (and have been observed elsewhere in the Python ecosystem), which could cause loss of sensitive clinical or proprietary commercial data. While our pilot security assessment pipeline produced no suspicious results, future researchers will have to find ways of reducing false positives, sifting more easily through the reams of data, and handling assessment of IP addresses.
This security assessment of open source bioinformatics packages should be viewed as preliminary and only a first step. Others should consider building upon this work to help ensure the future security of open source bioinformatics software. Please email jmeyers@iqt.org if you have further interest in this topic and would like to discuss this research or associated tools. You can also find the code used to do the security analysis here.
Further Reading
“Bewear! Python Typosquatting Is About More Than Typos,” IQT Blog, September 2020.
“Bioviz Under the Microscope,” Part 1 and 2, IQT Labs Human-Machine Interfaces Team Blog, February 2020.
Čarnogurský, Martin, “Attacks on Package Managers,” Bachelor’s Thesis, Masaryk University, May 2019.
Acknowledgements
Thank you to Luke Berndt, Michael Chadwick, Zigfried Hampel-Arias, and Adam Van Etten for thoughtful review and critique.

